Linear algebra¶
Linear algebra is a fast and efficient means of solving silent substitution problems.
Basic example¶
[51]:
from pysilsub.problems import SilentSubstitutionProblem as SSP
# Instantiate the problem class for a 5-primary system
ssp = SSP.from_package_data('BCGAR')
# Define problem
ssp.ignore = ['rh']
ssp.target = ['sc']
ssp.silence = ['mc', 'lc', 'mel']
ssp.background = [.5] * ssp.nprimaries
ssp.target_contrast = .3
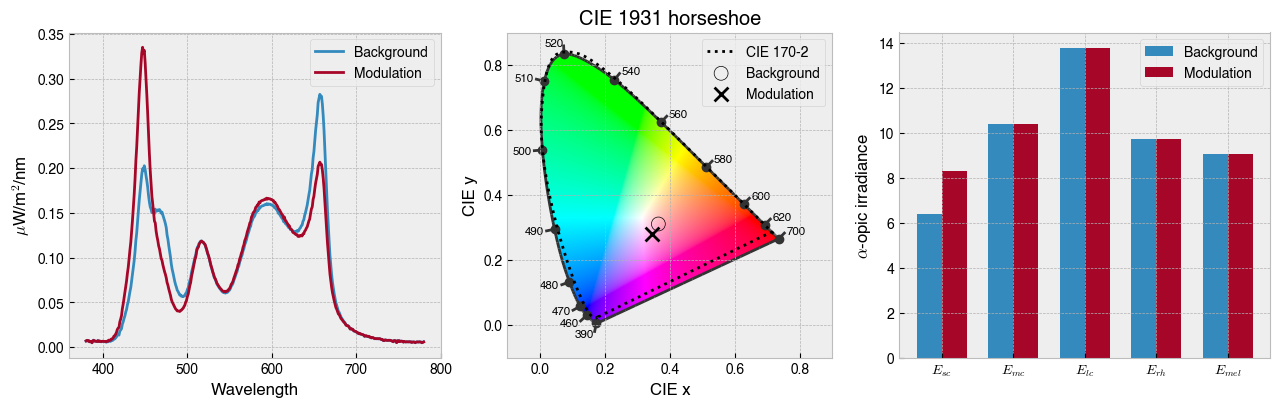
ssp.print_problem()
# Find solution
solution = ssp.linalg_solve()
# Plot solution
fig = ssp.plot_solution(solution)
# Show device settings
print(f'The background settings are: {ssp.w2s(ssp.background)}')
print(f'The modulation settings are: {ssp.w2s(solution)}')
************************************************************
*************** Silent Substitution Problem ****************
************************************************************
Device: BCGAR (8-bit, linear)
Observer: ColorimetricObserver(age=32, field_size=10)
Ignoring: ['rh']
Silencing: ['mc', 'lc', 'mel']
Targeting: ['sc']
Target contrast: [ 0.3]
Background: [0.5, 0.5, 0.5, 0.5, 0.5]
The background settings are: [127, 127, 127, 127, 127]
The modulation settings are: [245, 56, 129, 132, 83]
Worked example¶
Here’s what just happened.
For a background spectrum we take the mixture of all primaries at half-max power, \(\alpha_{bg} = [.5 \ .5 \ .5 \ .5 \ .5]\). We then use this to compute the matrix \(P_{bg}\), whose rows contain the spectral power distributions for each primary component of the background spectrum.
Note that in this case we are not summing the predicted spectra as we’ll be doing matrix algebra involving the individual primaries.
[112]:
a_bg = [.5, .5, .5, .5, .5]
P_bg = ssp.predict_multiprimary_spd(a_bg, nosum=True)
P_bg.T
[112]:
| Wavelength | 380 | 381 | 382 | 383 | 384 | 385 | 386 | 387 | 388 | 389 | ... | 771 | 772 | 773 | 774 | 775 | 776 | 777 | 778 | 779 | 780 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Primary | |||||||||||||||||||||
| 0 | 0.001098 | 0.001134 | 0.000991 | 0.001091 | 0.000879 | 0.000891 | 0.001021 | 0.000812 | 0.001010 | 0.001188 | ... | 0.000595 | 0.000620 | 0.000643 | 0.000634 | 0.000629 | 0.000625 | 0.000577 | 0.000516 | 0.000605 | 0.000600 |
| 1 | 0.000856 | 0.000933 | 0.000838 | 0.000828 | 0.000745 | 0.000710 | 0.000804 | 0.000523 | 0.000840 | 0.000927 | ... | 0.000465 | 0.000517 | 0.000513 | 0.000519 | 0.000556 | 0.000533 | 0.000525 | 0.000398 | 0.000539 | 0.000559 |
| 2 | 0.001115 | 0.001191 | 0.001198 | 0.001231 | 0.001003 | 0.000958 | 0.001071 | 0.000718 | 0.001226 | 0.001268 | ... | 0.000667 | 0.000768 | 0.000741 | 0.000678 | 0.000777 | 0.000710 | 0.000693 | 0.000562 | 0.000733 | 0.000826 |
| 3 | 0.001842 | 0.002067 | 0.001945 | 0.001945 | 0.001673 | 0.001608 | 0.001715 | 0.001290 | 0.001955 | 0.002015 | ... | 0.002439 | 0.002435 | 0.002404 | 0.002383 | 0.002361 | 0.002349 | 0.002305 | 0.002146 | 0.002114 | 0.002350 |
| 4 | 0.001736 | 0.002040 | 0.001812 | 0.002044 | 0.001556 | 0.001693 | 0.001622 | 0.001239 | 0.001834 | 0.002078 | ... | 0.001141 | 0.001277 | 0.001299 | 0.001251 | 0.001467 | 0.001355 | 0.001301 | 0.001093 | 0.001240 | 0.001354 |
5 rows × 401 columns
Next, we need to compute the matrix of \(a\)-opic irradiances:
\(A = P_{bg} \cdot S_{a}\)
where the columns of \(S_{a}\) are the photoreceptor action spectra of the observer.
[100]:
from pysilsub.CIE import get_CIES026_action_spectra
S_a = get_CIES026_action_spectra()
S_a
[100]:
| sc | mc | lc | rh | mel | |
|---|---|---|---|---|---|
| Wavelength | |||||
| 380 | 0.0 | 0.000000 | 0.000000 | 5.890000e-04 | 9.181600e-04 |
| 381 | 0.0 | 0.000000 | 0.000000 | 6.650000e-04 | 1.045600e-03 |
| 382 | 0.0 | 0.000000 | 0.000000 | 7.520000e-04 | 1.178600e-03 |
| 383 | 0.0 | 0.000000 | 0.000000 | 8.540000e-04 | 1.322800e-03 |
| 384 | 0.0 | 0.000000 | 0.000000 | 9.720000e-04 | 1.483800e-03 |
| ... | ... | ... | ... | ... | ... |
| 776 | 0.0 | 0.000002 | 0.000024 | 1.730000e-07 | 2.550000e-08 |
| 777 | 0.0 | 0.000002 | 0.000023 | 1.640000e-07 | 2.420000e-08 |
| 778 | 0.0 | 0.000002 | 0.000021 | 1.550000e-07 | 2.290000e-08 |
| 779 | 0.0 | 0.000002 | 0.000020 | 1.470000e-07 | 2.170000e-08 |
| 780 | 0.0 | 0.000001 | 0.000019 | 1.390000e-07 | 2.050000e-08 |
401 rows × 5 columns
The dot product of these matrices gives \(A\)
[106]:
A = S_a.T.dot(P_bg)
A
[106]:
| Primary | 0 | 1 | 2 | 3 | 4 |
|---|---|---|---|---|---|
| sc | 3.580009 | 2.453689 | 0.211358 | 0.095264 | 0.057067 |
| mc | 0.627573 | 1.355843 | 3.573256 | 4.683028 | 0.156777 |
| lc | 0.435727 | 0.873636 | 2.915142 | 8.686473 | 0.853310 |
| rh | 1.730730 | 2.965132 | 3.638927 | 1.288265 | 0.076642 |
| mel | 2.094721 | 3.520715 | 2.841600 | 0.509312 | 0.069365 |
Now, given a set of scaling coefficients for the primaries, \(\alpha_{sc}=\left[p_1\ p_{2\ }p_{3\ }p_{4\ }p_5\right]\), the forward model predicts that the output modulation \(\beta=\left[sc\ mc\ lc\ rh\ mel\right]\) is \(\beta= \alpha_{sc}A\).
To invert this process, we calculate \(\alpha_{sc}=\beta A^{-1}\), which yields the desired scaling coefficients that must be added to the primary weights for the background in order to produce the desired output modulation \(\beta\).
[113]:
import numpy as np
import pandas as pd
# Calculate inverse of A
A1 = pd.DataFrame(
np.linalg.inv(A.values),
A.columns,
A.index)
A1
[113]:
| sc | mc | lc | rh | mel | |
|---|---|---|---|---|---|
| Primary | |||||
| 0 | 0.473797 | -0.159332 | -0.001803 | 0.993080 | -1.104764 |
| 1 | -0.279487 | 0.340021 | -0.037472 | -1.654925 | 1.750941 |
| 2 | -0.002424 | -0.286409 | 0.015329 | 1.342032 | -1.022070 |
| 3 | 0.026572 | 0.502356 | -0.059770 | -0.855885 | 0.523675 |
| 4 | -0.218010 | -4.402157 | 1.767262 | 5.315177 | -3.067724 |
[114]:
# Requested modulation for S-cones
requested_contrast = .2
# sc, mc, lc, rh, mel
b = np.array([requested_contrast, 0., 0., 0., 0.])
# Scale requested contrasts to percentage of background units
b = A.sum(axis=1).mul(b)
# Calculate the scaling coefficients
a_sc = A1.dot(b) / 2
a_sc
[114]:
Primary
0 0.303106
1 -0.178799
2 -0.001551
3 0.016999
4 -0.139469
dtype: float64
Following on from above, \(\alpha_{mod} = \alpha_{bg} + \alpha_{sc}\).
[115]:
a_mod = (a_bg + a_sc).to_list()
a_mod
[115]:
[0.8031061259025292,
0.32120125937804056,
0.4984491198072997,
0.5169994543850375,
0.36053050404907117]
Obviously, these values need to be between zero and one for the solution to be valid, which in this case they are. As before, we can visualise the solution.
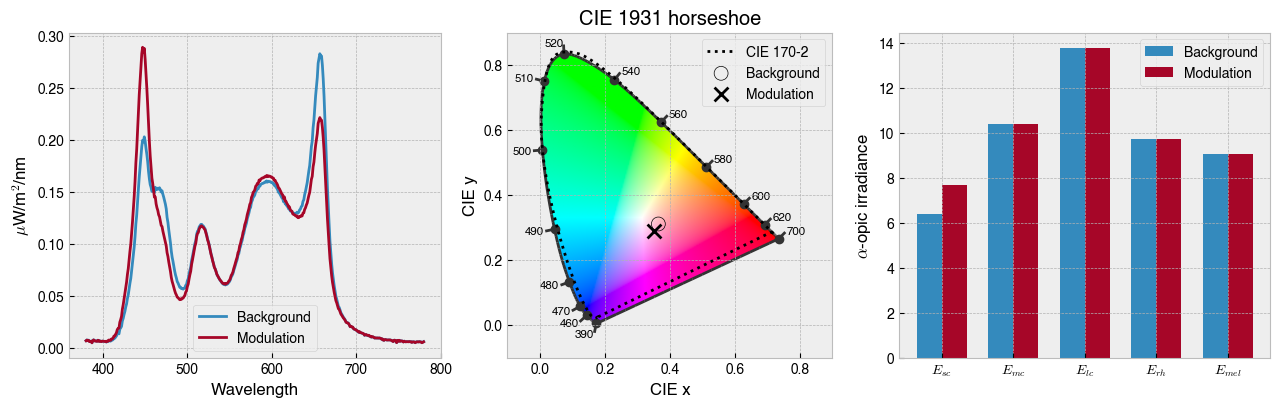
[116]:
# Plot the solutution
result_fig = ssp.plot_solution(a_mod)
# Show device settings
print(f'The background settings are: {ssp.w2s(a_bg)}')
print(f'The modulation settings are: {ssp.w2s(a_mod)}')
The background settings are: [127, 127, 127, 127, 127]
The modulation settings are: [204, 81, 127, 131, 91]
Contrast modulations¶
Multiprimary devices with good linearity, sufficient bit depth and rapid spectral switching capabilities may be used to present temporal modulations of photoreceptor-targeted contrast. Such stimuli have helped to shed light on how the pupil responds to input from different photoreceptors as a function of time (e.g., Barrionuevo et al., 2016; Spitschan et al., 2014).
To make contrast modulations, first define a stimulus profile with a sampling frequency in line with the switching capability of the stimulation device. In this case, its a 1 Hz sinusoid with 50 samples per cycle.
[117]:
import matplotlib.pyplot as plt
from pysilsub import waves
duration = 1 # s
frequency = 1 # Hz
sampling_frequency = 50 # Hz
stimulus_profile = waves.make_stimulus_waveform(
frequency=frequency,
duration=duration,
sampling_frequency=sampling_frequency
)
time = np.linspace(0, duration, len(stimulus_profile))
plt.plot(time, stimulus_profile)
[117]:
[<matplotlib.lines.Line2D at 0x7fd7c2fc0d30>]
Now, define a problem. Here we define the background spectrum as all primaries at half-max power so we can maximise bidirectional contrast. Then we say that we want to ignore rods, minimize contrast on M-cones, L-cones and melanopsin, and modulate the S-cones.
[118]:
# Load some example data for a 10-primary system
ssp = SSP.from_package_data('STLAB_1_York')
ssp.background = [.5] * ssp.nprimaries
ssp.ignore = ['rh']
ssp.silence = ['mc', 'lc', 'mel']
ssp.target = ['sc']
ssp.print_problem()
************************************************************
*************** Silent Substitution Problem ****************
************************************************************
Device: STLAB_1 (binocular, left eye)
Observer: ColorimetricObserver(age=32, field_size=10)
Ignoring: ['rh']
Silencing: ['mc', 'lc', 'mel']
Targeting: ['sc']
Target contrast: None
Background: [0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5, 0.5]
Next, choose a peak contrast value that is known to be in gamut and solve for each point in the stimulus profile. The algebraic approach is well suited here as it guarantees a linear uni-directional scaling of the primaries and is less computationally expensive than optimisation.
[119]:
peak_contrast = .3 # Known to be within gamut
solutions = []
for point in stimulus_profile:
ssp.target_contrast = point * peak_contrast
solutions.append(ssp.linalg_solve())
# Plot the modulation
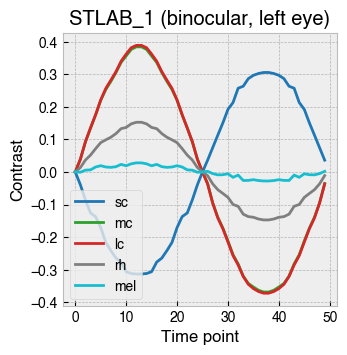
_ = ssp.plot_contrast_modulation(solutions)
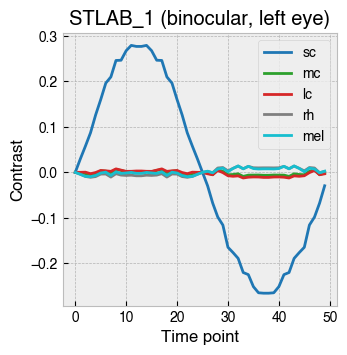
Plotting the forward projection of contrast for each solution in this case reveals a smooth S-cone modulation with little contrast splatter on the other photoreceptors.
Finally, convert the solutions to settings compatible with the native resolution of the stimulation device.
[120]:
device_settings = [ssp.w2s(s) for s in solutions]
device_settings = pd.DataFrame(device_settings, index=time)
device_settings.index.name = 'time'
with pd.option_context('display.max_rows', 10):
print(device_settings)
0 1 2 3 4 5 6 7 8 9
time
0.000000 2047 2047 2047 2047 2047 2047 2047 2047 2047 2047
0.020408 2194 2194 2021 1977 1924 2033 2118 2034 1971 2014
0.040816 2338 2339 1995 1907 1804 2019 2189 2020 1897 1982
0.061224 2477 2480 1971 1840 1687 2006 2257 2008 1824 1951
0.081633 2610 2613 1947 1776 1576 1993 2322 1996 1756 1921
... ... ... ... ... ... ... ... ... ... ...
0.918367 1360 1356 2169 2377 2622 2113 1712 2110 2402 2201
0.938776 1484 1481 2147 2318 2518 2101 1772 2098 2338 2173
0.959184 1617 1614 2123 2254 2407 2088 1837 2086 2270 2143
0.979592 1756 1755 2099 2187 2290 2075 1905 2074 2197 2112
1.000000 1900 1900 2073 2117 2170 2061 1976 2060 2123 2080
[50 rows x 10 columns]
Now you just need to tell the stimulation device to set the primary inputs to these values at the specified timepoints and it will produce the S-cone modulation shown above.
Note that with this method one can also modulate multiple photoreceptors together.
[121]:
ssp.ignore = ['rh']
ssp.silence = ['sc', 'mel']
ssp.target = ['mc', 'lc']
peak_contrast = .2 # Known to be within gamut
solutions = []
for point in stimulus_profile:
ssp.target_contrast = point * peak_contrast
solutions.append(ssp.linalg_solve())
# Plot the modulation
_ = ssp.plot_contrast_modulation(solutions)
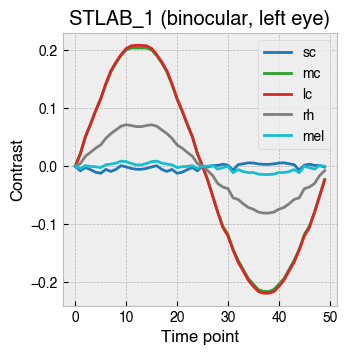
We can even modulate photoreceptors in counterphase.
[122]:
ssp.ignore = ['rh']
ssp.silence = ['mel']
ssp.target = ['sc', 'mc', 'lc']
peak_contrast = .35 # Known to be within gamut
solutions = []
for point in stimulus_profile:
c = point * peak_contrast
ssp.target_contrast = [-c, c, c]
solutions.append(ssp.linalg_solve())
# Plot the modulation
_ = ssp.plot_contrast_modulation(solutions)